If your CUT&Tag experiment failed or if you have very low yields (< 30 ng total DNA), there are several areas of the CUTANA™ CUT&Tag workflow that may require troubleshooting. Below we provide a generalized CUT&Tag troubleshooting strategy, followed by more detailed considerations for customized troubleshooting. Note that each approaches relies on the incorporation of EpiCypher-recommended control antibodies (H3K4me3, H3K27me3, and IgG) and the SNAP-CUTANA™ K-MetStat Panel, as well as our comprehensive control checks (Figure 1).
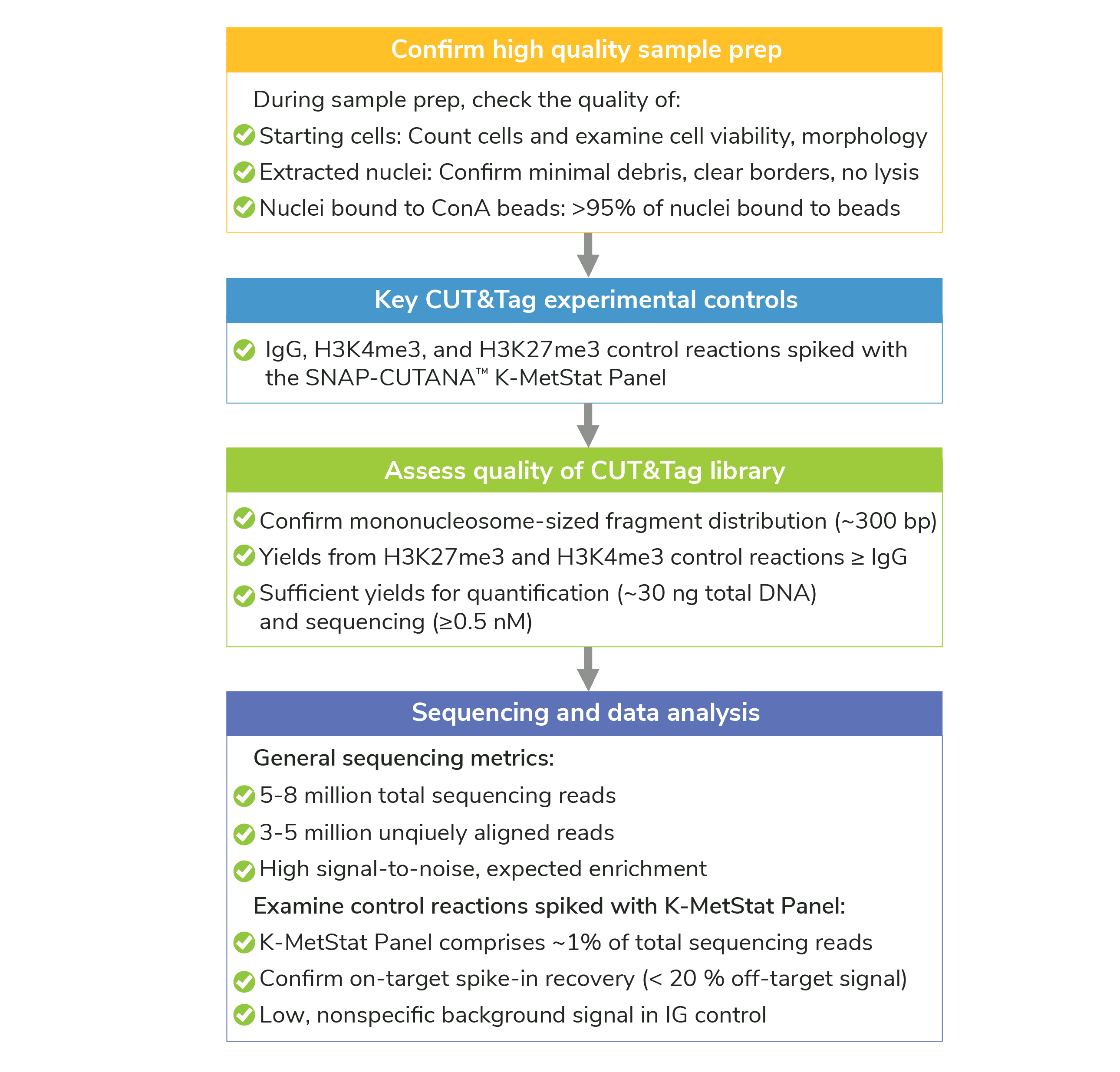
Figure 1: Summary of controls and success metrics for CUT&Tag experiments.
Troubleshooting Summary
Repeat the experiment using 100,000 native nuclei per reaction. Make the following adjustments:
Include reactions with H3K4me3, H3K27me3, and IgG control antibodies spiked with the K-MetStat Panel.
Test multiple antibodies to experimental target(s).
Test adjustments to nuclei extraction as outlined below.
Confirm sample prep quality, avoid ConA bead loss, and optimize PCR cycling conditions.
Mix reactions with a nutator and resuspend beads per the CUTANA CUT&Tag protocol.
Examine results:
If controls work but experimental targets fail: Confirm target is correctly localized to chromatin (e.g. check stimulation conditions); test additional antibodies and/or cross-linking conditions.
If controls AND experimental targets show no yield: Try purifying DNA prior to PCR using CUTANATM DNA Purification Beads (EpiCypher 21-1407) or a column-based DNA purification kit.
If yields are still <0.5 nM despite extensive optimization, try increasing the number of indexing PCR cycles, which will improve yields for fragment distribution analysis and enable pooling at standard concentrations for sequencing. Note that this strategy may increase read duplication rates. Deeper sequencing is recommended to capture library diversity.
If it is not possible to repeat the experiment, use a Speedvac to increase library concentration and add as much of the library as possible to the sequencing pool. Deeper sequencing is recommended to ensure sufficient read depth and fully capture library diversity.
Sample prep troubleshooting
Sample prep is one of the main areas we help troubleshoot for CUT&Tag experiments, and we have developed comprehensive quality control checks (Figure 1) to ensure robust nuclei extraction and ConA bead binding.
Questions to consider
What is your cell/sample type? See Sample Prep section for specific recommendations.
Have you confirmed sample prep and ConA bead binding, per the quality control steps outlined in Figure 1? Are final extracted nuclei of high quality (not lysing)?
Are you using the recommended 100,000 nuclei per reaction? If using low nuclei numbers, remember that CUT&Tag success from low inputs depends on antibody quality and target abundance. See the Antibody Validation section of this site.
Does the TapeStation trace show strong enrichment of primer-dimers? This often occurs when samples are of low quality, or when the target has low abundance.
Confirm proper ConA bead storage conditions and quality. Are ConA beads brown and stored at 4˚C? ConA beads should NEVER be frozen.
Are both EpiCypher-recommended control antibodies and antibodies to experimental targets failing? This may indicate an issue with sample prep.
Nuclei Lysis and Bead Clumping:
If you are experiencing issues with nuclei lysis or bead clumping during CUT&Tag Sample Prep, consider the following adjustments:
Reduce nuclei extraction incubation time; monitor lysis under a microscope to optimize.
Resuspend the extracted nuclei pellet in 1XPBS or Wash Buffer 1, instead of Nuclei Extraction Buffer. The resuspension volume can be increased if desired.
Perform ConA bead binding in PBS or Wash Buffer 1, instead of Nuclei Extraction Buffer.
Antibodies and control reactions in troubleshooting
High quality antibodies are critical to robust CUT&Tag profiles. We include multiple CUT&Tag-validated control antibodies in our workflows to ensure reliable experimental workflows and assay monitoring.
Questions to consider
Have you included reactions with control antibodies and the K-MetStat Panel? These controls are crucial for troubleshooting CUT&Tag.
Is your antibody validated for CUT&Tag? Antibodies that are robust in other applications, such as ChIP-seq, may perform poorly in CUT&Tag. See the Antibody Validation section of this site for guidance.
Have EpiCypher-recommended control antibodies worked, but antibodies to experimental targets failed? This indicates that your antibody is of poor quality or is incompatible with CUT&Tag.
Are you trying to map transcription factors or other chromatin proteins using CUT&Tag? We recommend using CUT&RUN for mapping chromatin proteins, as CUT&Tag is most robust for histone PTMs.
Troubleshooting recommendations
Include control reactions spiked with the K-MetStat Panel in every experiment. If the control antibodies generate robust peaks and expected spike-in data, you can be confident in your workflow.
If control antibodies work as expected but experimental targets continue to fail:
Test additional antibodies to your histone PTM target. See the Antibody Validation section of this site, or contact us for specific recommendations.
Test cross-linking vs. native conditions.
Confirm target is correctly localized to chromatin. For instance, if you are mapping histone phosphorylation in response to immunostimulation, make sure your cellular stimulation is successful.
Additional CUT&Tag workflow troubleshooting
In addition to sample prep, antibody validation, and control reactions, the CUT&Tag workflow can also require troubleshooting.
Questions to consider
Have reactions been mixed properly using a nutator? A nutator gently rocks tubes back and forth, which is recommended for CUT&Tag. Avoid rotating tubes, which results in beads dry out and reduces yields.
Have ConA beads become clumpy or dried out during the protocol? It is important to keep beads in suspension, particularly during pAG-Tn5 binding and tagmentation.
Are the indexing PCR parameters correct? Check the CUTANA CUT&Tag Protocol to make sure.
Are both EpiCypher-recommended control antibodies and antibodies to experimental targets failing? This could be a sample prep issue, but may also be related to fundamental problems with the CUT&Tag workflow.
Troubleshooting recommendations
Remake all buffers on the first day of the experiment. Check sample quality as outlined above.
Be sure to use a nutator to mix samples and follow reaction mixing instructions as outlined in the CUTANA CUT&Tag protocol.
Increase the number of PCR cycles to improve yields for sequencing; this strategy may increase read duplicates and requires deeper sequencing to capture library diversity
Try purifying DNA prior to PCR using CUTANATM DNA Purification Beads or a column-based DNA purification kit.