SNAP-CUTANA™ Spike-ins are a defined control added to immobilized cells or nuclei at the start of CUTANA™ CUT&RUN and CUT&Tag chromatin mapping experiments. They are specifically designed for reactions targeting histone post translational modifications (PTMs), such as histone lysine methylation and epitope tagged-proteins (HA and DYKDDDDK), and allow users to examine antibody performance, workflow success, and much more. Because SNAP-CUTANA Spike-ins replicate chromatin structure (i.e. nucleosomes), the natural target of CUTANA assays, they provide accurate on- and off-target substrates for antibody specificity testing. When paired with highly validated control antibodies, they can also be used as a readout for CUTANA assay success.
Features and Advantages
Fast: SNAP-CUTANA Spike-ins are coupled to magnetic beads, allowing them to be added to CUTANA reactions in one quick step for seamless workflow integration (see the SNAP-CUTANA K-MetStat Panel, Figure 1).
Easy: No protocol modifications are necessary. SNAP-CUTANA Spike-in data are quickly analyzed in final sequencing data using our step-by-step instructions.
Reliable: Use these controls to examine sample quality, MNase activity, antibody specificity, and troubleshoot challenging workflows.
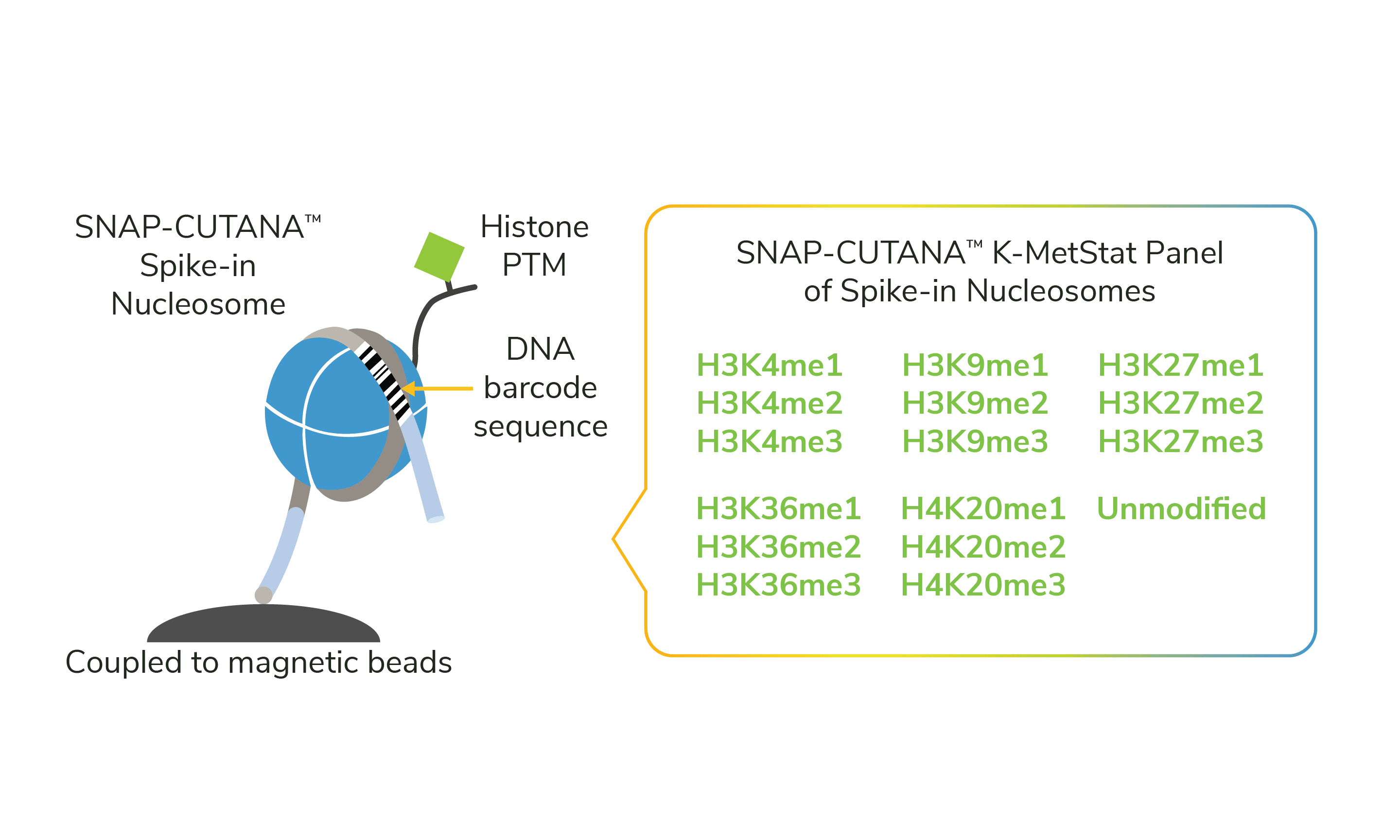
Figure 1. SNAP-CUTANA Spike-in Controls are pools of highly pure nucleosomes carrying defined histone PTMs. For instance, the SNAP-CUTANA K-MetStat Panel shown in this figure is a pool of 16 nucleosomes representing 16 distinct methyl-lysine states. Spike-in nucleosomes are individually coupled to magnetic beads and pooled into a single panel for convenient one-step addition to CUTANA workflows. pAG-MNase (CUT&RUN) or pAG-Tn5 (CUT&Tag) cleavage releases all antibody-bound targets into solution, including antibody-bound spike-ins. Each spike-in nucleosome contains a PTM-specific DNA barcode sequence, which enables detection of K-MetStat Panel controls in sequencing data.
How do SNAP-CUTANA Spike-ins work in CUTANA assays?
You can add SNAP-CUTANA Spike-in Controls to CUT&RUN and CUT&Tag reactions that are mapping target included in the Panel. SNAP-CUTANA Spike-ins are added to reactions just prior to addition of target-specific antibody (Figure 2).
Immobilization of spike-in nucleosomes on magnetic beads makes them similar to bead-coupled cells, allowing both to be captured using magnetic separation racks. This means that spike-ins can be added early in the experiment for side-by-side processing with bead-coupled cells, all in the same reaction tube. Spike-ins can thus report on multiple aspects of CUTANA workflows, including antibody specificity and pAG-MNase (CUT&RUN) or pAG-Tn5 (CUT&Tag) activity.
.png)
Figure 2. Schematic showing how SNAP-CUTANA Spike-ins, such as the K-MetStat Panel, are used during CUTANA workflows. *SNAP-CUTANA Spike-ins can also be added to experimental reactions mapping a target in the panel.
No other modifications to the protocol are required. You simply continue your CUTANA workflow, adding your target specific antibody for overnight incubation. Antibodies bind their specific histone PTM target in permeabilized cells AND in the SNAP-CUTANA Spike-in Panel. The next day, pAG-MNase or pAG-Tn5 is used to cleave antibody-bound chromatin (in cells) and antibody-bound spike-ins (from beads). DNA fragments from the clipped spike-ins are processed alongside sample DNA for library prep and sequencing.
The PTM-specific DNA barcode sequences on the spike-in nucleosomes (Figures 1 and 2) are used to detect reads from the spike-ins vs. cells in sequencing data. Generally, we aim for ~1% of total sequencing reads to be from SNAP-CUTANA Spike-ins, although this may vary based on sequencing depth and target abundance. The main goal is to have many thousands of sequencing reads aligned to SNAP-CUTANA Spike-ins, which enables adequate sampling of the panel and reliable use in downstream applications.
When assessing the reads assigned to spike-ins, you should see enrichment for on-target PTM and minimal reads from off-target PTMs in the Panel (Figure 2). These data can be used as a direct readout for PTM recovery and assay success, described in more detail here. For a more detailed explanation about our epitope-tag SNAP-CUTANA Spike-ins, see this blog.