At EpiCypher, we have developed two main applications of the SNAP-CUTANA™ Spike-in Controls for CUTANA CUT&RUN and CUT&Tag assays. The first is determination of workflow success, discussed here. The other application of SNAP-CUTANA Spike-ins is for histone PTM antibody validation.
Why test histone PTM antibodies?
Histone PTM antibodies are notoriously nonspecific, which can result in misleading biological interpretations. EpiCypher is well aware of this problem. We first developed SNAP nucleosome spike-in technology for ChIP-seq (SNAP-ChIP™ Spike-ins) and used them to test more than 400 lysine-methylation and lysine-acylation antibodies in ChIP assays. We found that >70% of histone PTM antibodies show significant off-target binding and/or low binding efficiency – even highly cited antibodies! The results of this study can be found at chromatinantibodies.com.
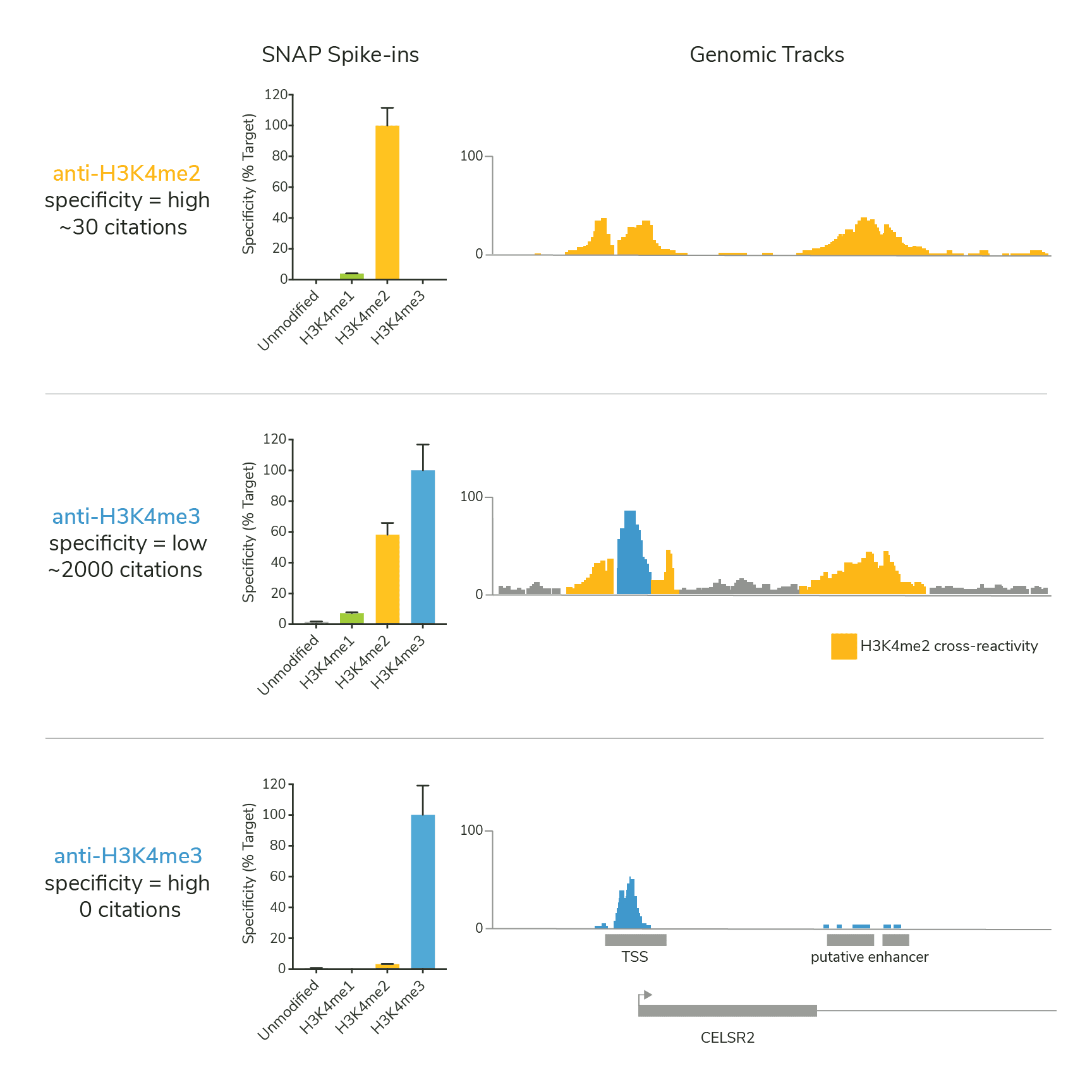
Figure 1. A highly cited H3K4me3 antibody fails SNAP Spike-in testing and shows extensive cross-reactivity to H3K4me2 (middle panel). Highly specific H3K4me2 (top panel; orange) and H3K4me3 (bottom panel; blue) antibodies are shown for comparison. See Shah et al. 2018.
These results were a major driving factor in the development of SNAP-CUTANA Spike-in technology. To ensure the success of emerging CUTANA™ epigenomic assays, defined controls are needed to find the best antibodies possible.
Why use SNAP-CUTANA Spike-ins vs. other antibody validation methods?
This strategy is the only method that directly confirms antibody specificity in CUTANA assays against physiological on- and off-target substrates. EpiCypher knows from experience that histone PTM antibodies do not perform the same across assays – for instance, a good ChIP antibody is NOT guaranteed to work in CUTANA assays!
Metrics for SNAP-CUTANA antibody validation in CUTANA assays
EpiCypher is using these defined nucleosome spike-in controls to identify best-in-class histone PTM antibodies for CUTANA assays. We offer a collection of SNAP-Certified Antibodies for CUT&RUN and can provide antibody recommendations upon request.
We validate antibody specificity as well as efficiency, allowing users to be confident when using reduced cell numbers. Each of our SNAP-Certified Antibodies show:
High specificity: <20% recovery of off-target PTMs
High target efficiency: Robust profiling at 500,000 and 50,000 starting cells
How to use SNAP-CUTANA Spike-ins for PTM antibody testing
Purchase a SNAP-CUTANA Panel that contains your histone PTM target. For instance, you can use the K-MetStat Panel for any reaction that is mapping a histone methyl-lysine mark included in the panel.
Source several antibodies to your histone PTM target to test, ideally from several sources and/or targeting distinct epitopes.
Test antibodies side-by-side in CUTANA assays using the recommended amount of cells or nuclei in each reaction.
Add the SNAP-CUTANA Spike-in Panel to CUTANA reactions just before addition of primary antibody, as indicated in the protocol. Include control reactions with the K-MetStat Panel to confirm workflow success. See this article for an overview.
Aim for <20% antibody cross-reactivity and consistent genomic enrichment with 500,000 cells (or 100,000 nuclei for CUT&Tag).
In CUT&RUN, to help ensure high antibody efficiency, validation at 50,000 cells is also recommended.
Additional notes
Do NOT add a SNAP-CUTANA Panel to reactions targeting a protein or a PTM that is not represented in the panel.
Note that for CUTANA CUT&RUN Kit users, this will require purchase of additional K-MetStat Panel.