The SNAP-CUTANA™ Spike-ins replicate physiological chromatin and carry defined on- and off-target histone PTMs, making them the ideal control to report on CUT&RUN and CUT&Tag assay success. In CUTANA assay workflows, EpiCypher adds the SNAP-CUTANA™ K-MetStat Panel to positive (H3K4me3, H3K27me3) and negative (IgG) control reactions to confirm workflows and guide troubleshooting.
Why are these controls useful?
SNAP-CUTANA Spike-in controls are panels of defined nucleosomes, making them the only control that replicates the in vivo target of chromatin mapping assays. The spike-ins are added to cells immobilized on ConA beads and are processed alongside the sample throughout the CUTANA workflow (Figure 1). By combining these spike-ins with our rigorously validated CUTANA antibodies for H3K4me3, H3K27me3, and IgG, we can immediately gauge workflow success.
EpiCypher includes these control reactions in every experiment, and it has saved us countless time and resources in protocol optimization. Should you reach out to our technical support team for troubleshooting assistance, they will be able to assist you much better if you include data derived from this panel. For details on how to leverage the K-MetStat Panel for troubleshooting, see this article.
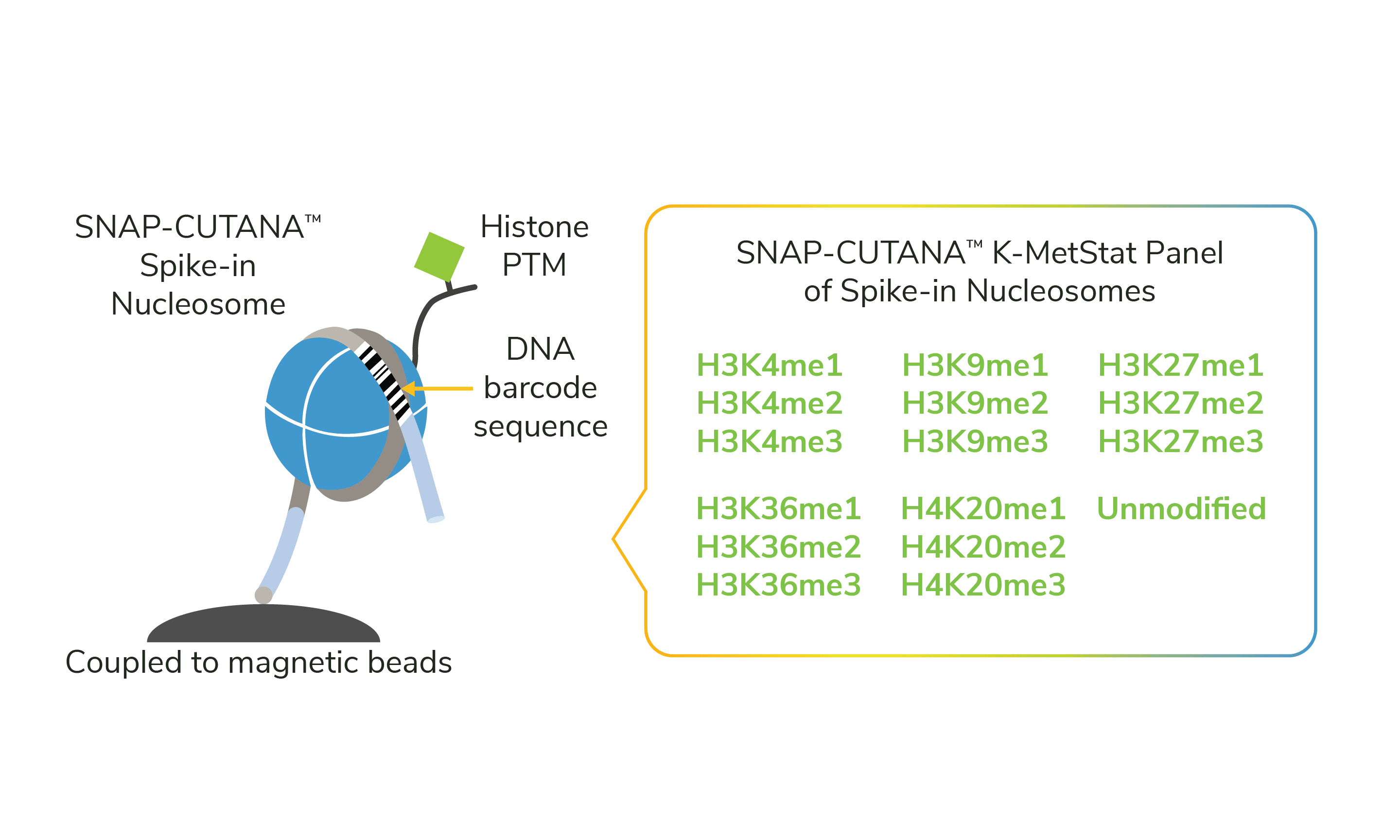
Figure 1. The K-MetStat Panel is composed of 16 spike-in nucleosomes, representing 15 distinct histone lysine methylation PTMs and an unmodified control. Nucleosomes are coupled to magnetic beads for easy one-step addition to CUT&RUN. A PTM-specific DNA barcode enables detection of spike-ins in sequencing data.
Pairing the K-MetStat Panel with control antibodies to determine assay success
We review the specifics of how SNAP-CUTANA Spike-ins are processed in CUTANA assays here; the strategy using the K-MetStat Panel with control antibodies is outlined in Figure 2. Note that Figure 2 is applicable for CUT&RUN, CUT&Tag, and related immunotethering assays. Briefly:
Add the SNAP-CUTANA K-MetStat Panel to designated control reactions immediately prior to the addition of H3K4me3, H3K27me3, or IgG control antibody (Figure 2).
Add antibody, which binds its target in cells and in the spike-in panel (Figure 2).
The CUTANA enzyme, pAG-MNase (CUT&RN) or pAG-TN5 (CUT&Tag), cleaves antibody-bound chromatin and antibody-bound spike-in. Cleaved DNA is purified and prepared for sequencing according to the respective workflow.
Samples are sequenced. For each control reaction, download sequencing files and determine the number of sequencing reads aligned to each PTM-specific DNA barcode, using the analysis procedure as described in this article. Barcode read counts provide a useful measurement of PTM recovery and workflow success.
.png)
Figure 2. The K-MetStat Panel is used with control antibodies to determine CUTANA assay success, as outlined below. Two different outcomes are shown. *Note that the K-MetStat Panel can be added to experimental reactions targeting a PTM in the Panel. Additional K-MetStat Panel can be purchased at epicypher.com/19-1002.
What are the expected results? How is assay success determined?
Figure 2 illustrates two potential outcomes from H3K4me3, H3K27me3, and IgG control reactions: one successful CUTANA experiment and one that indicates a failed experiment. In each heatmap, reactions are separated into rows and recovery of each PTM in the Panel is separated by column. The heatmap shows data normalized to the on-target PTM (i.e. the on-target PTM is set to 100%, appearing blue). Off-target PTMs should have low signal relative to this target and appear orange. EpiCypher always aims for off-target PTMs to show <20% signal relative to the target PTM.
To determine assay success, keep the following metrics in mind:
The IgG negative control should show no preference among PTMs and low background.
H3K4me3 and H3K27me3 positive control reactions should have strong enrichment their respective on-target spike-in. Both reactions should show less than 20% off-target PTM recovery and high signal-to-noise.
Spike-in barcode reads should comprise ~1% (0.5-5%) of total sequencing reads. This may vary based on target abundance and sequencing depth. The main goal is to have thousands of reads, which will allow adequate sampling of the K-MetStat Panel for reliable data analysis.
If control reactions generate expected spike-in data, you can be confident in the technical aspects of your workflow (Figure 2, left heatmap).
If more than 20% off-target PTM recovery in H3K4me3 control and/or high background in IgG control indicate experimental problems (Figure 2, right heatmap). See this article for a discussion of troubleshooting using spike-in results.