Library fragments at ~150 bp are adapter dimers, which result from self-ligation of sequencing adapters and preferential amplification during library prep (see Figure 1). As long as you see predominant enrichment of mononucleosome-sized fragments at ~300 bp, and adapter dimers comprise less than 5% of your prepared library, you can move forward with sequencing.
If you have >5% adapter dimers in your library, we recommend removing them using CUTANATM DNA Purification Beads (EpiCypher 21-1407), following ratio recommendations on the “Application Notes” tab. Note that we suggest pooling sequencing libraries and then removing adapter dimers, as opposed to processing individual libraries.
If adapter dimers are the only peak in your fragment distribution trace, see we suggest repeating the experiment with more cells, and be sure to perform quality control checks for sample prep.
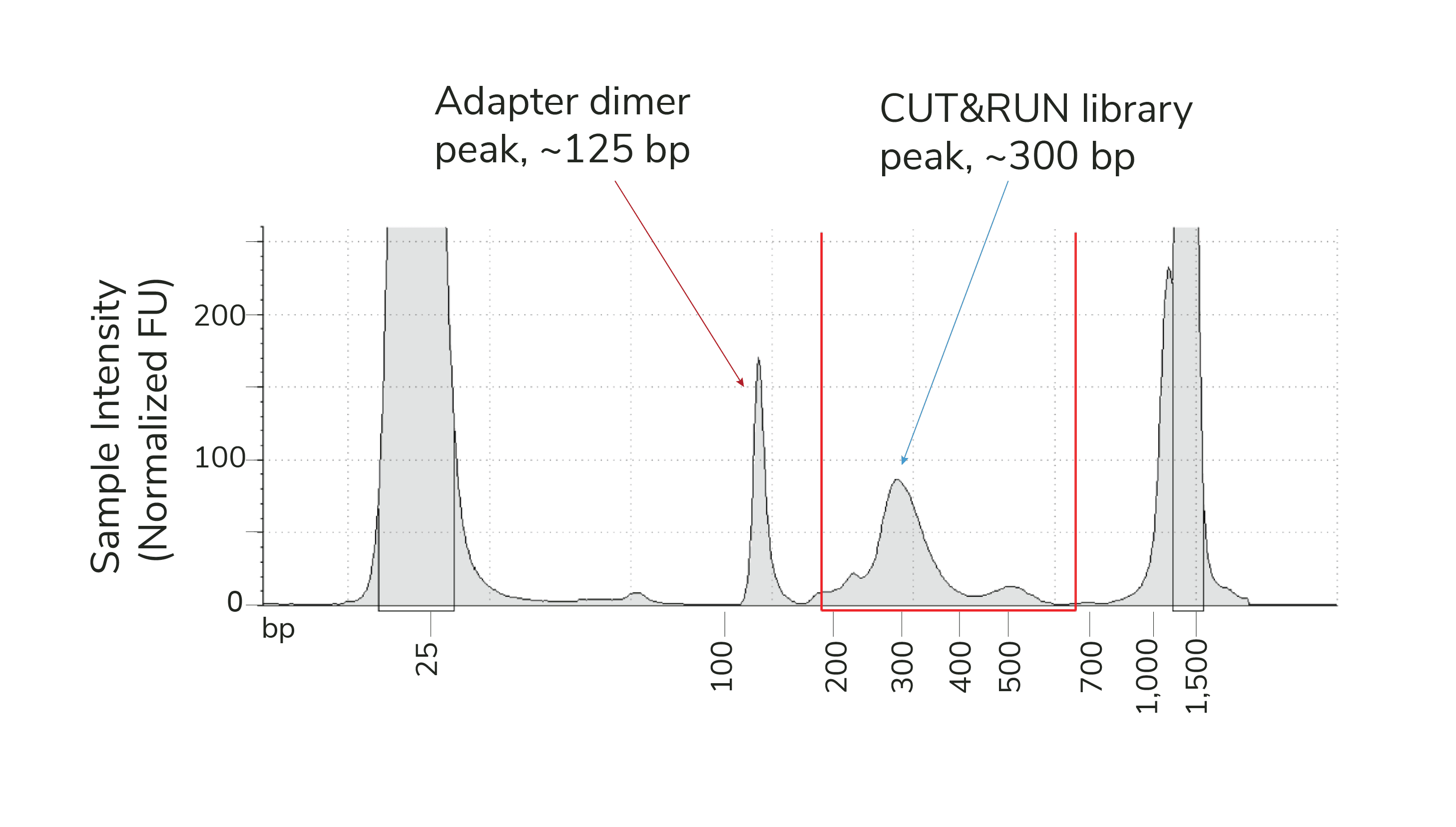
Figure 1. Example TapeStation trace from CUT&RUN H3K27me3 library containing an adapter dimer peak (~150 bp, red arrow) and expected library peak (~300 bp, blue arrow). Red lines denote the 200-700 bp range, used to determine library concentration.