Primer dimers are generated by self-ligation of sequencing primers that are preferentially amplified due to their small size. Primer dimer contamination appears as a peak at ~100 bp (Figure 1), and is caused by bad antibodies, low nuclei numbers, poor tagmentation efficiency, and/or indexing PCR setup.
If you observe significant primer dimers (>5% of the library), repeat the experiment using 100,000 native nuclei per reaction. Include reactions with H3K4me3, H3K27me3, and IgG control antibodies spiked with the SNAP-CUTANA™ K-MetStat Panel. If controls work but experimental targets fail, test additional antibodies as described in the Antibody Validation section of this site.
Alternatively, you may consider using the Sequencing Library Cleanup protocol in our CUTANA™ Quick Cleanup DNA Purification Kit manual. Note that removal of primer dimers is performed on the entire multiplexed sequencing pool, NOT individual libraries, to minimize sample loss.
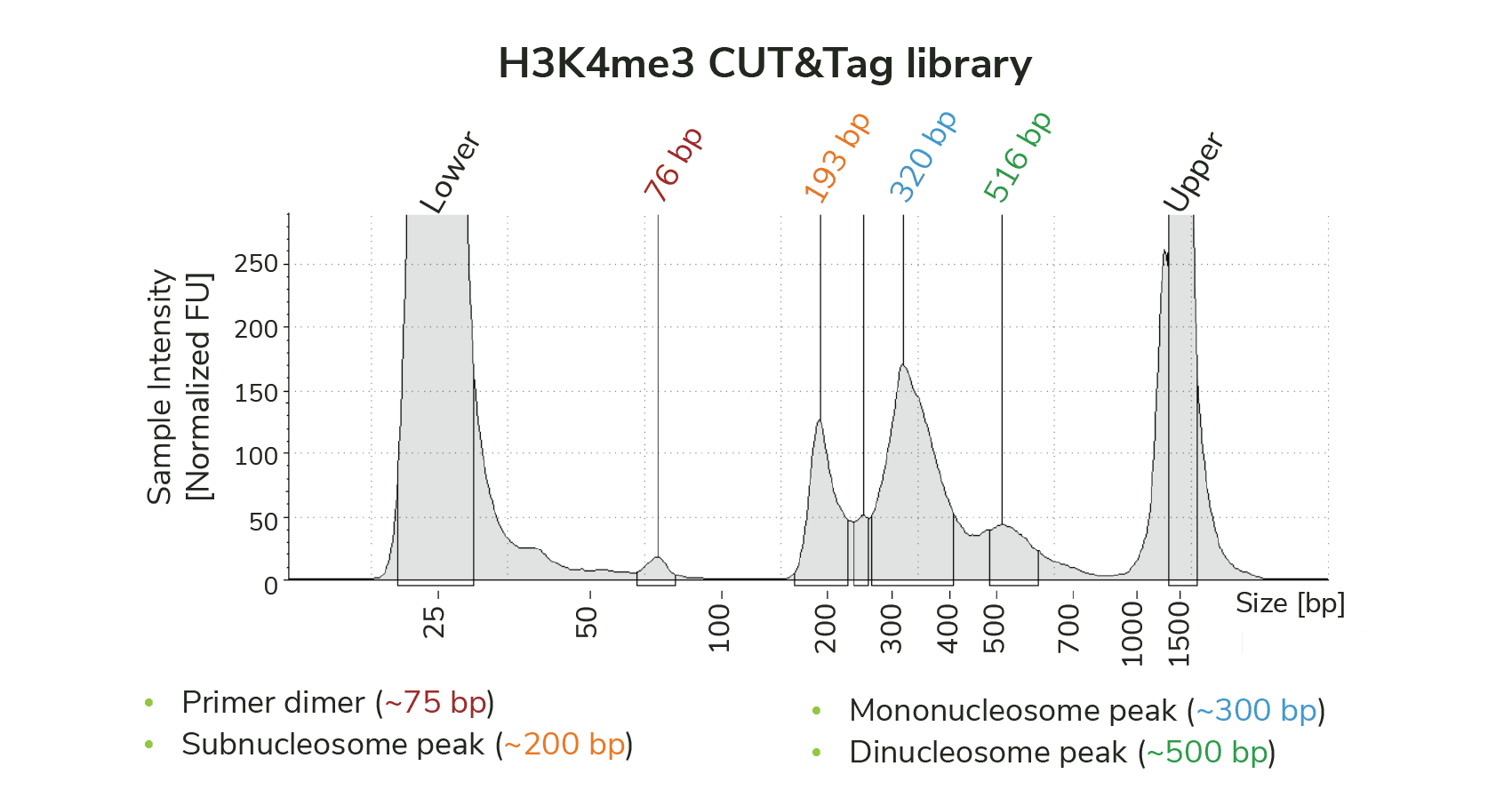
Figure 1: TapeStation trace for CUTANA CUT&Tag library targeting H3K4me3 shows a small amount of primer dimer contamination (~75 bp). However, the library is primarily enriched for mononucleosome-sized fragments (~300 bp), and does not require additional cleanup prior to sequencing.