Fragments between 25 and 100 bp are likely primer dimers (Figure 1), which occur when indexing PCR primers preferentially self-ligate. Enrichment of primer dimers often indicates problems with tagmentation efficiency and/or PCR amplification, and are most common when performing CUT&Tag with low numbers of nuclei.
If primer dimers comprise >5% of the library, additional cleanup should be performed prior to sequencing. We recommend performing cleanup on pooled libraries, not individual libraries, to minimize sample loss. Use the bead:DNA ratios suggested in the Application Notes for our CUTANA™ DNA Purification Beads (EpiCypher 21-1407) for maximal recovery.
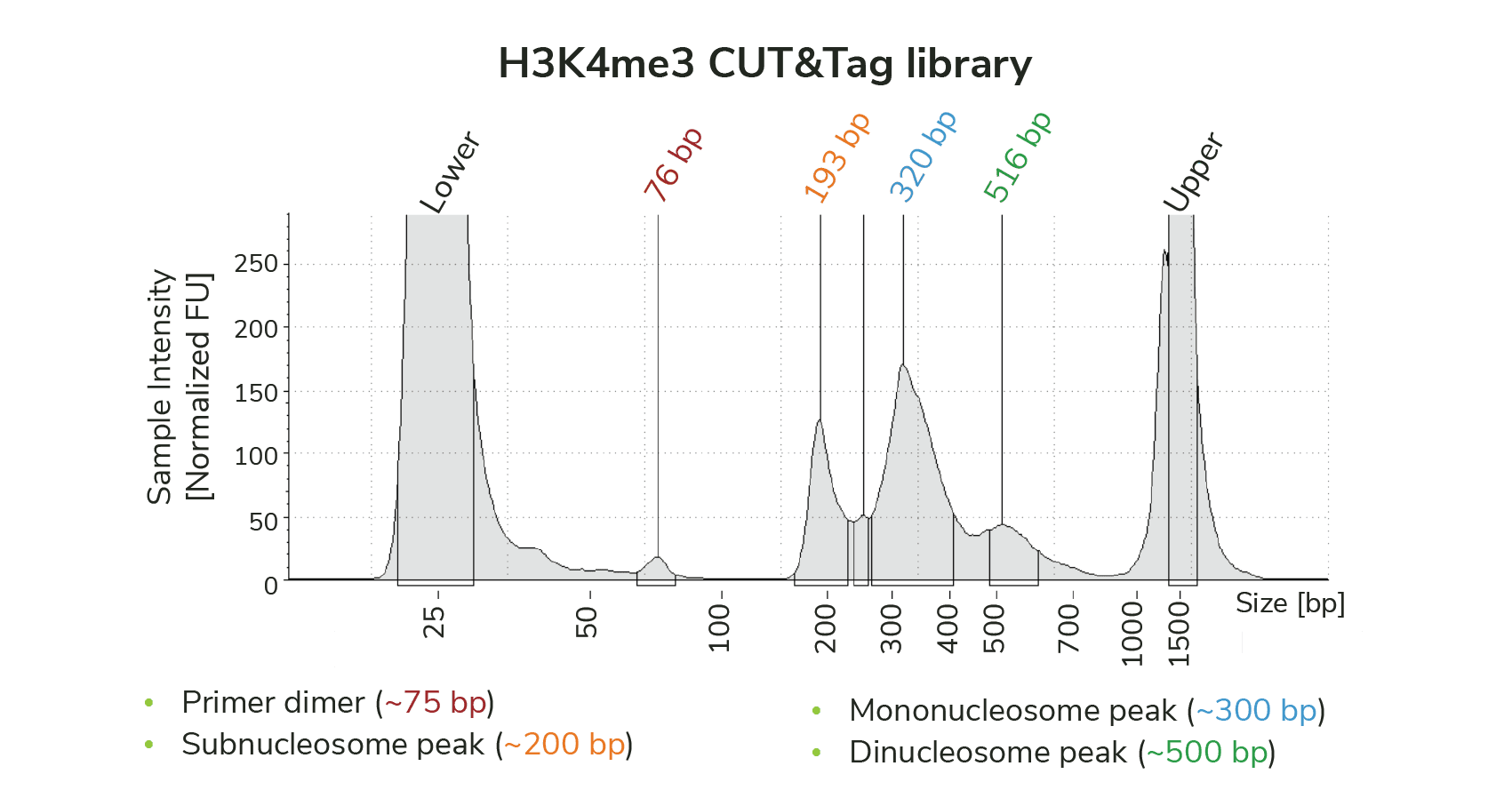
Figure 1: TapeStation trace for CUTANA CUT&Tag library targeting H3K4me3 shows a small amount of primer dimer contamination (~75 bp). However, the library is primarily enriched for mononucleosome-sized fragments (~300 bp), and does not require additional cleanup prior to sequencing.