Important Notes
If using intact cells for CUT&RUN or CUT&Tag, the concentration of Digitonin must be optimized for efficient cell permeabilization. Insufficient Digitonin prevents antibody and enzyme from entering the cell, while excess amounts may result in cell lysis. EpiCypher recommends using the minimal amount of Digitonin required to permeabilize >95% of cells. Optimize Digitonin concentrations for each cell type used in CUT&RUN as outlined below.
If using nuclei for CUT&RUN or CUT&Tag, Digitonin optimization is NOT required. 0.01% Digitonin is added to assay buffers to prevent the beads from forming a film on the side of tubes.
Label tubes
Label five fresh 1.5 mL tubes with percent Digitonin (see Table 1) and a sixth tube as Control.
Label 6 additional tubes with percent Digitonin or as Control. This second set of tubes will be used for cells.
Prepare buffers
Prepare a series of five Cell Permeabilization Buffers using 5% Digitonin and CUT&RUN Wash Buffer (or CUT&Tag Wash Buffer 1). Prepare buffers FRESH on the day of use. Add the appropriate volume of Wash Buffer to each tube as outlined in the Table 1. Add 10 µL 5% Digitonin to the first tube, labeled 0.05%. Vortex to mix.
Prepare the other four Cell Permeabilization Buffers by serial dilution (see Table 1). Vortex each buffer to mix and place on ice.
For the Control buffer, prepare 0.05% DMSO in Wash Buffer.
Final % Digitonin | 0.05 | 0.01 | 0.001 | 0.0001 | 0.00001 |
|---|---|---|---|---|---|
Volume from previous tube (µL) | - | 200 | 100 | 100 | 100 |
Wash buffer (µL) | 990 | 800 | 900 | 900 | 900 |
5% Digitonin (µL) | 10 | - | - | - | - |
Table 1: Use to prepare serial dilutions of Digitonin.
Permeabilize cells
Harvest cultured cells for permeabilization testing. To determine the number of cells needed for Digitonin optimization, multiply the number of cells used per reaction x 6.2 (six tubes + 20% excess volume for pipetting errors).
CUT&RUN: 500,000 cells x 6.2 = 3.1 million cells
CUT&Tag: 100,000 cells x 6.2 = 620,000 cells
Spin 600 x g, 3 min, room temperature (RT). Remove supernatant. Resuspend cells in 620 µL RT 1X PBS.
Aliquot 100 µL cells to the second set of labeled tubes that were set aside for cells.
Spin cells at 600 x g for 3 min at room temperature (RT). Remove supernatant. Resuspend each cell pellet in 100 µL of the assigned Permeabilization Buffer (or Control) and incubate 10 minutes at RT.
At the end of the incubation, examine each sample by Trypan Blue staining. Count live (intact, Trypan negative) vs. dead (permeabilized, Trypan positive) cells. Select minimum Digitonin concentration that permeabilizes >95% of cells (example in Figure 1).
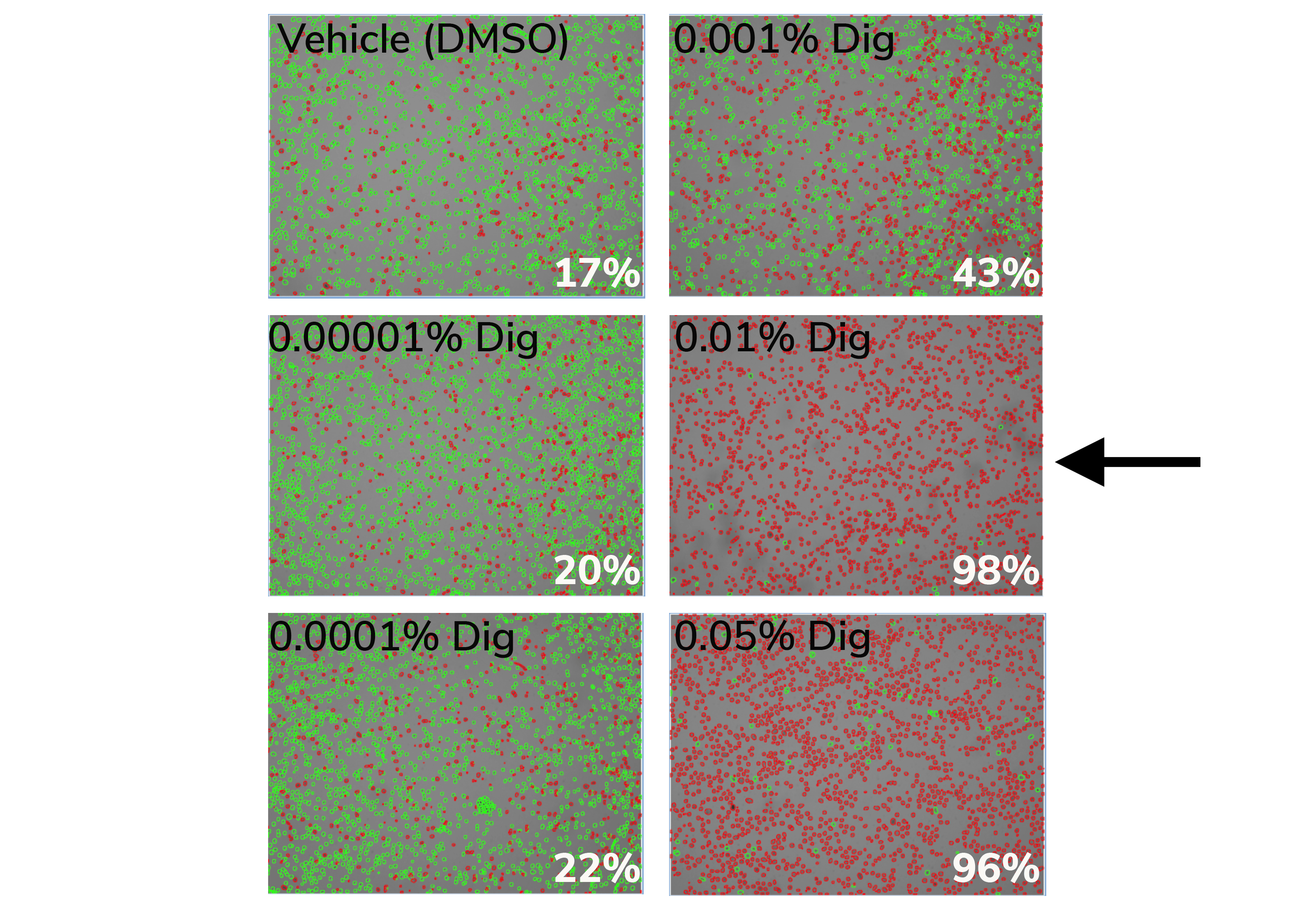
Figure 1. 0.01% Digitonin is the minimum concentration that permeabilizes >95% K562 cells (black arrow). Cells were treated with CUT&RUN Wash Buffer containing various Digitonin concentrations and evaluated by Trypan Blue staining. Green cells (Trypan negative) are intact, whereas permeabilized/dead cells (Trypan positive) are red. Values (bottom right of each panel) indicate percent of dead/permeabilized cells.