There are two general approaches to confirming the quality of your CUT&RUN sequencing library, outlined below.
Library yields
In general, library yields should not be used to determine assay success. Library yields vary widely by cell type, number of cells, target abundance, and antibody quality.
Aim for a library concentration of ≥ 1 nM, which will enable pooling at standard concentrations for multiplexed sequencing.
For library concentrations below 0.5 nM, see see this article.
Fragment distribution of purified library
Fragment distribution analysis of purified sequencing libraries is the single BEST method to confirm CUT&RUN success.
Libraries should show enrichment of mononucleosome-sized DNA fragments at ~300 bp, which includes CUT&RUN DNA + sequencing adapters (see Figure 1).
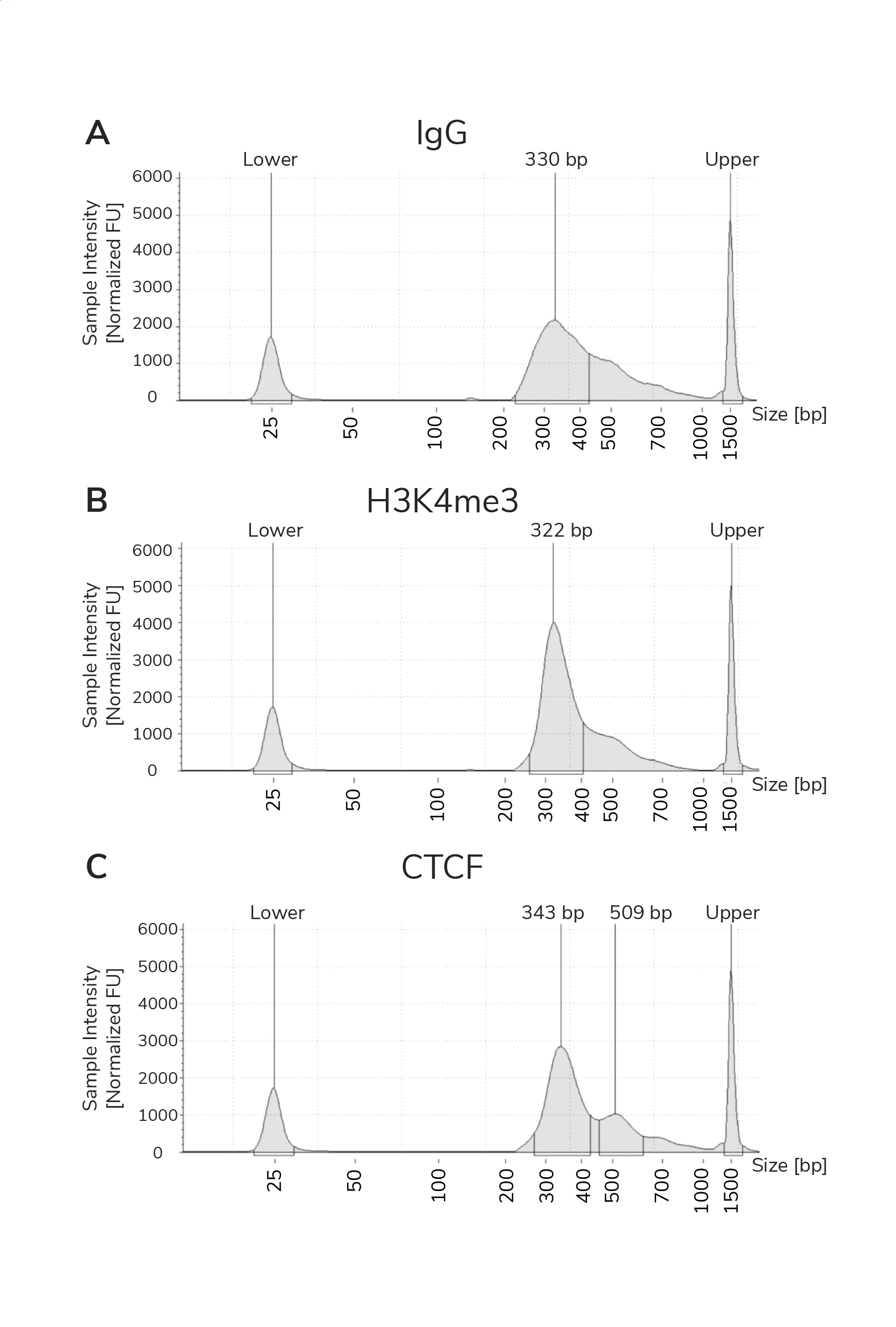
Figure 1. Typical TapeStation traces from CUTANA™ CUT&RUN libraries prepared using antibodies targeting IgG (EpiCypher 13-0042), H3K4me3 (EpiCypher 13-0060), and CTCF (EpiCypher 13-2014). All libraries are predominantly enriched for mononucleosome-sized fragments, as indicated by the peak at ~300 bp (~170 bp nucleosomes + sequencing adapters).