High background in CUT&Tag sequencing data could indicate poor sample prep, over-digestion by Tn5, DNA damage, incorrect PCR cycling conditions, or antibody failure.
In troubleshooting experiments, use 100,000 native nuclei per reaction. Include reactions with H3K4me3, H3K27me3, and IgG control antibodies spiked with the SNAP-CUTANA™ K-MetStat Panel. Test multiple antibodies to experimental targets. Pay close attention to the following:
Confirm sample prep by assessing the quality of your sample at three steps:
At initial cell harvest, prior to nuclei extraction: Do cells have >80% viability and expected morphology? Is there unexpected lysis or debris?
After nuclei extraction: Are nuclei round, and detached from cell components? Is there visible lysis or large amounts of debris in sample?
After binding to ConA beads: Ensure nuclei have bound to beads, and check that little to no nuclei are in the supernatant.
Follow instructions in the manual for mixing CUT&Tag reactions at key steps. For instance, during pAG-Tn5 binding and activation, it is important to keep ConA beads resuspended. Likewise, during final CUT&Tag library cleanup, it is essential to thoroughly mix the sample with SPRI beads to ensure efficient binding.
Double check that indexing PCR parameters are accurate.
Confirm that TapeStation/Bioanalyzer traces are enriched for mononucleosome-sized fragments at ~300 bp.
Examine sequencing data from positive and negative control reactions:
Are expected peaks generated (Figure 1)?
Are appropriate K-MetStat Spike-ins recovered (see this article)?
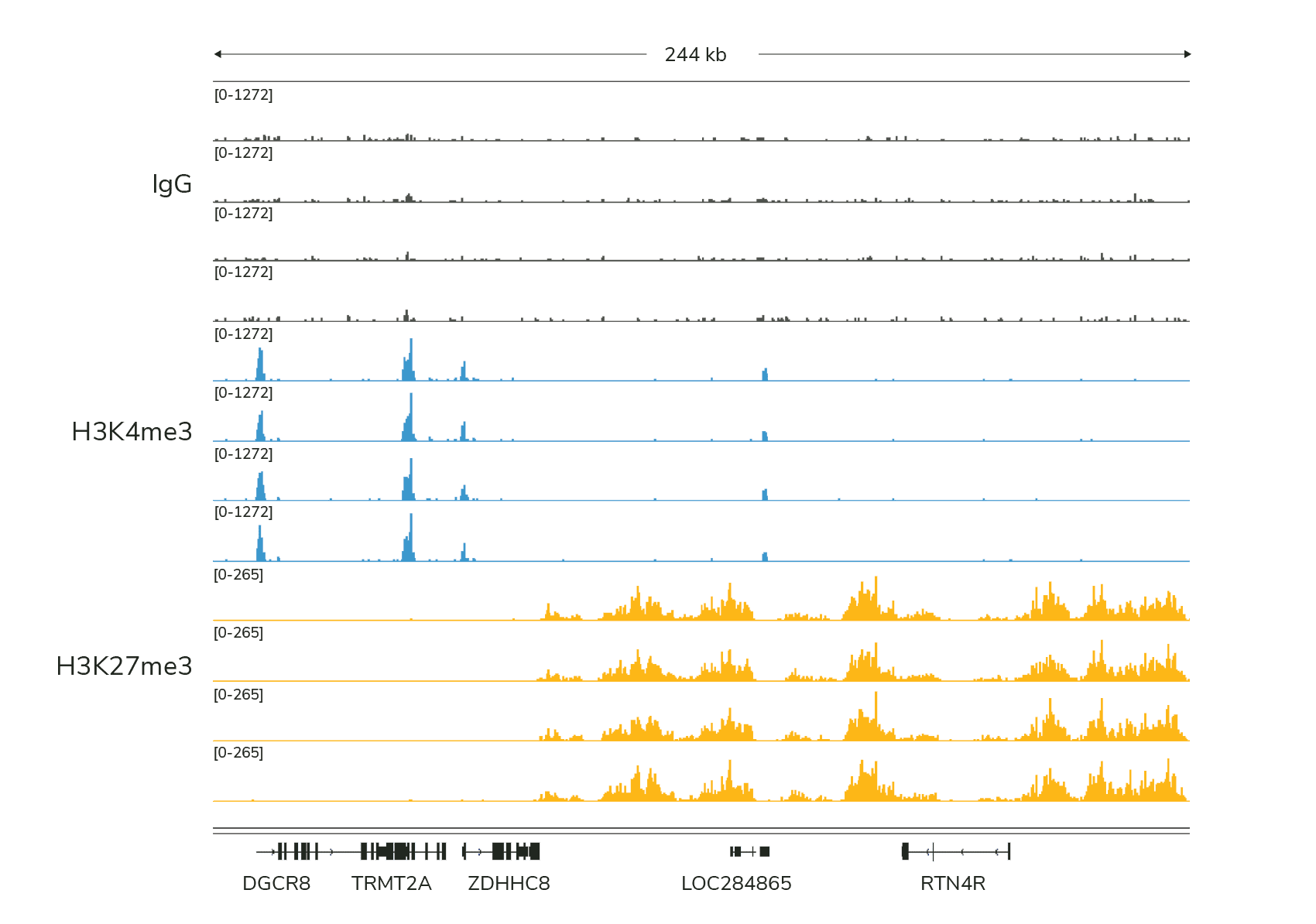
Figure 1: Typical sequencing results generated using IgG, H3K4me3, and H3K27me3 control antibodies in CUTANA™ CUT&Tag experiments. Reactions were performed using 100,000 K562 nuclei.