There are two general approaches to confirm the quality of CUT&Tag sequencing libraries:
Fragment distribution of purified library
Fragment distribution analysis of purified sequencing libraries is the single BEST method to confirm CUT&Tag success. Libraries should show enrichment of mononucleosome-sized DNA fragments (~300 bp, including CUT&Tag DNA + sequencing adapters).
Use the H3K4me3 and H3K27me3 positive control reactions and IgG negative control reactions (Figure 1) to help confirm CUT&Tag success.
The "trident pattern" is observed for many histone PTM CUT&Tag libraries, including the H3K27me3 and H3K4me3 libraries in Figure 1B & C. These fragments are generated during tagmentation and subsequent indexing PCR. Peaks represent:
Subnucleosome fragments: ~200 bp, orange text in Figure 1. Note that these fragments are NOT adapter dimer/primer dimer.
Mononucleosome fragments: ~300 bp, blue text in Figure 1. Predominant enrichment at ~300 bp is the best metric for CUT&Tag success.
Dinucleosome fragments: ~500 bp, green text in Figure 1.
Primer dimer fragments: ~25-100 bp, red text in Figure 1. If primer dimers comprise >5% of the library, additional cleanup should be performed; see our recommendations for bead:DNA ratios to use in the Application Notes for our CUTANA™ DNA Purification Beads (EpiCypher 21-1407).
The trident peak pattern is not cause for concern, as it typically reflects on-target enrichment. They do not impact sequencing and we do NOT recommend removing them, as it risks loss of the mononucleosome peak (~300 bp).
Library yields
There is no typical yield for CUT&Tag. Library yields vary widely by cell type, number of cells, target abundance, and antibody quality.
In general, aim for 2 ng/μL or ~30 ng total DNA, which will allow accurate library quantification and minimize PCR duplicates. Library molarity >0.5 nM for the 200-700 bp region will allow pooling at standard concentrations for sequencing.
For library concentrations below 0.5 nM, see the CUT&Tag Optimization and Troubleshooting section of this website for guidance.
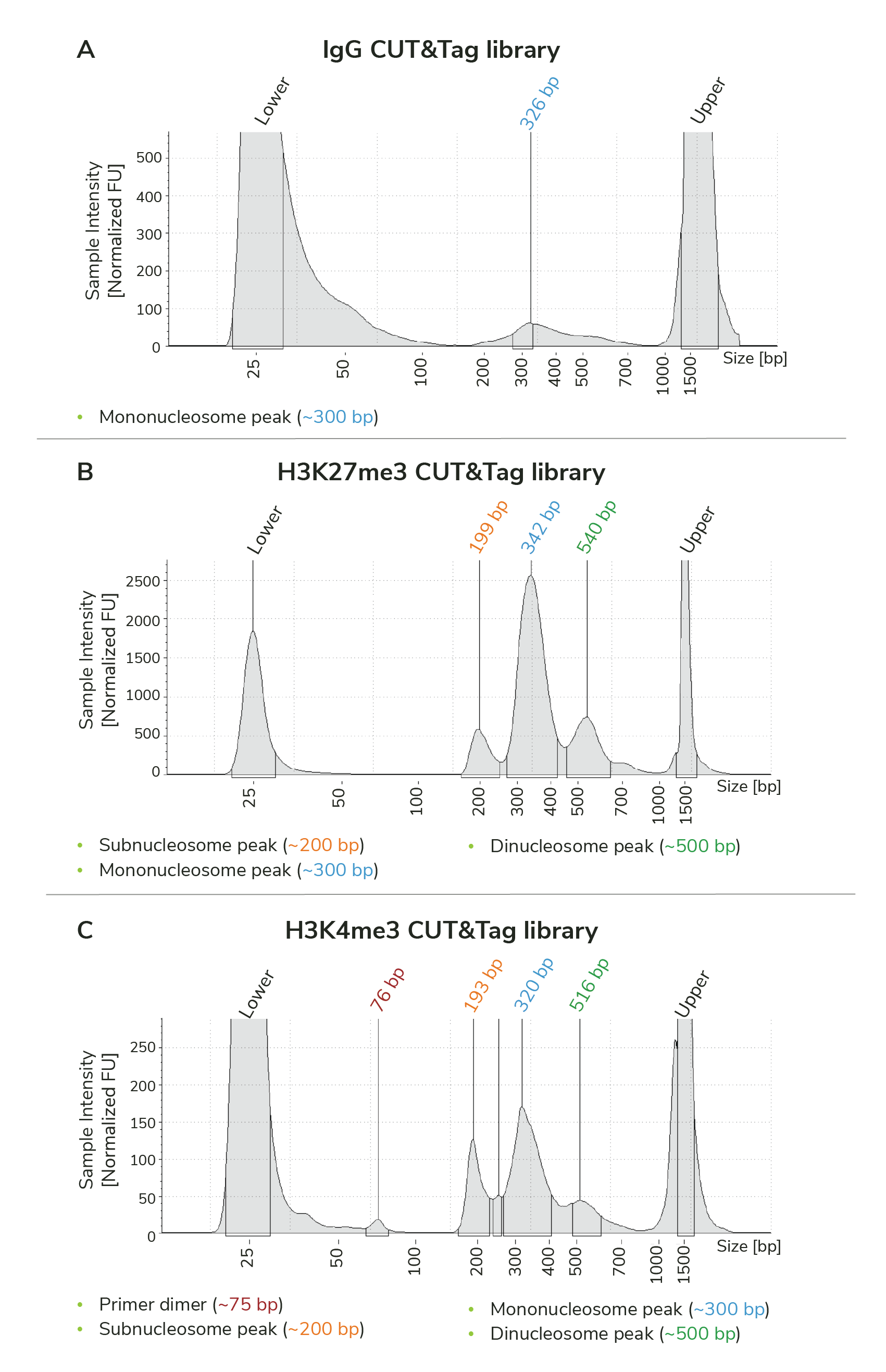
Figure 1. Typical TapeStation traces from CUTANA CUT&Tag libraries targeting IgG (negative control), H3K27me3 (EpiCypher 13-0055), and H3K4me3 (EpiCypher 13-0041 shown; replaced by 13-0060). All libraries are predominantly enriched for mononucleosome-sized library fragments, indicated by the peak at ~300 bp (blue text). Subnucleosome (~200 bp; orange text) and dinucleosome (~500 bp; green text) peaks are present, forming the trident patten discussed above. Minimal primer dimers are observed in the H3K4me3 library (~75 bp; red text).